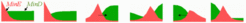A minimum mechanism to generate oscillating polar patterns
The use of highly dynamic patterns generated by a destabilization antagonist is widespread. On other pages it has been shown that this mechanism is appropriate to explain the pigmentation pattern of the shells of mollusks , phyllotaxis and the orientation of chemotactic cells and growth cones by small external gradients.
In fields small compared with the range of the antagonistic reaction polar patterns emerge. If such a polar pattern becomes destabilized by a second long-lasting antagonistic reaction, the polar pattern starts to oscillate. In the simulation shown below activator maxima form at the pole but disappear after some time due to the slowly accumulating second inhibitor (pink). A new activation emerges at the previously non-activated side (which is not poisoned by the long-lasting inhibitor) (equation and parameters ).
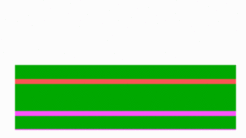
Although the out-of-phase oscillation was correctly described, it was clear that this mechanism was incompatible with the known components. There was no indication for an inhibitor. On the other hand, in the simulation shown above, there is no role for a more centrally localized component such as MinE. Without MinE, no oscillation takes place and MinD is stable and homogeneously attached to the membrane. How can a more centrally located component be responsible for the flashing up at the poles?
Modeling has revealed that these pattern-forming reactions depend on interaction of the activator-depleted substrate type . The self-enhancing process consists in the assembly at the membrane. Bound molecules enhance the binding of more molecules. This goes on the expense of unbound molecules that can rapidly diffuse in the cytoplasm. Thus, the different ranges for the self-enhancing and the antagonistic component are straightforward in such an interaction. The simulation below shows the formation of a local maximum (red) on the expense of diffusible precursor molecules (grey).

In the subsequent simulation it is assumed that the MinE maximum depends on a component sitting at the membrane (MinD, green) that is removed by the maximum. The maximum start to move into regions with more MinD. The direction reverses after reaching the other pole.