A more static mechanism for center detection:
Insertion of new maxima at maximum distance from the poles
In the Gram positive rod, B. subtilis, MinE is absent and no pole-to-pole oscillations of MinC/MinD occur. Instead, stable maxima of this division inhibitor are maintained at both poles . Under their repelling influence, the Z-ring assembles at the largest possible distance from both maxima, i.e., at the center [1-3]. Although a detailed modeling for B. subtilis has to be worked out, the general models suggest a relatively simple mechanism that allows for center-finding in a more static system.
If pattern forming reactions are at work, new maxima are formed during growth. This can happen either by a split and shift of existing maxima, or by the trigger of new maxima in the enlarging interstices. Splitting is the prevalent mode if the antagonistic reaction is accomplished by the depletion of a substance required for autocatalysis, and has been discussed above. In contrast, if an activator /inhibitor reaction is involved, new maxima become inserted whenever the distance to existing maxima exceeds a certain value. At these positions, the inhibitor (red) concentration becomes so low that the onset of autocatalysis can no longer be suppressed. In the first simulation, a small but growing field is assumed (equation and parameters). Initially a polar pattern emerges. If a certain size is surpassed, a second maximum is generated at the opposite pole. The result is a symmetrical pattern:
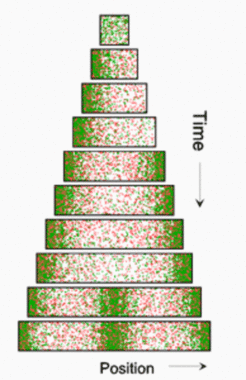


Upon further growth, the inhibitor concentration in the center decreases until a new activation is triggered here. If this new maximum induces the separation into two daughter cells, the two daughter cells would have again a symmetrical pattern with a maximum at each pole, and so on.
Again, when using such a mechanism for center-finding in a bacterium, the circumferential extension is too large to be neglected. The signal at the center must have the geometry of a stripe, not of a patch. According to the model, a saturation of the self-enhancement leads to activator distributions that have a long extension in one dimension but only a short extension in the other. Without initial asymmetries, these stripes will have somewhat random distributions. As shown in the simulation below, the inhibitory influence from the poles would lead automatically to an orientation of this stripe perpendicular to the long axis of the bacterium.

The rightmost simulation demonstrates that without saturation (see equation and parameters ), but otherwise identical parameters, the mechanism would generate isolated patches of activator accumulation.
References
- Edwards, D. H., and Errington, J. (1997). The Bacillussubtilis DivIVA protein targets to the division septum and controls the site specificity of cell division., Molec Microbiol 24, 905-915.
- Marston, A. L., and Errington, J. (1999). Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC., Mol Microbiol 33, 84-96.
- Marston, A. L., Thomaides, H. B., Edwards, D. H., Sharpe, M. E., and Errington, J. (1998). Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site., Genes & Dev 12, 3419-3430.



