Pole-to-pole oscillation of the division inhibitor MinC in E.coli : More details and time-lapse fluorescence micrographs
The preparation for division starts with the assembly of a polymeric ring of the tubulin-like GTPase FtsZ (Z-ring). This ring is localized to the center by the actions of the MinC, MinD, and MinE proteins. MinC inhibits the initiation of the Z-ring. MinC co-localizes with MinD. In wild type cells (WT), MinC/D forms a polar pattern that oscillates between the poles, keeping the center free for initiation of cell division. Thus, virtually all of MinC/D dynamically assembles on the membrane in the shape of a test-tube covering the membrane from one pole up to approximately midcell. Most of MinE accumulates at the rim of this tube, in the shape of a ring (the E-ring). The rim of the MinC/D tube and associated E-ring move from a central position to the cell pole until both the tube and ring vanish. Meanwhile, a new MinC/D tube and associated E-ring form in the opposite cell half and the process repeats, resulting in a pole-to-pole oscillation cycle of the division inhibitor. A full cycle takes about 50s. The panel below shows a schematic drawing of the MinC/D (green) and MinE (red) localization cycles.

Time-lapse fluorescence micrographs showing the dynamic behavior of Gfp-MinC. MinC is a division inhibitor which associates, and co-oscillates, with MinD. Pole-to-pole oscillation of MinD, in turn, requires the activity of MinE. On time average, the center of the cell is exposed to the lowest level of division inhibitor. Times are indicated in seconds. The last panel shows the cell viewed with DIC (differential interference contrast) optics. The bar in this panel represents 2 µm. Shown is a cell of strain PB114(lDB175)/pDR175DminCDE(Plac::minDE)/PlR::gfp-minC, cI857].
Cells were grown at 37oC in minimal medium supplemented with 50mM IPTG, and displayed a normal division pattern. For other details, see [1,2]
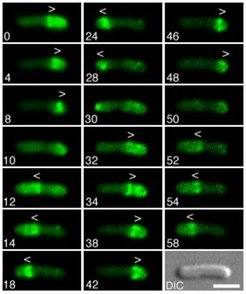
Time-lapse fluorescence micrographs showing the dynamic behavior of MinE-Gfp in normally dividing cells. Times are indicated in seconds. Note the accumulation of MinE-Gfp in the shape of a ring (the E-ring) as well as in an extra-annular peripheral pattern (PEA signal) which is present in between the ring and one of the cell poles. Note the net movement of a ring towards the pole with the PEA signal (0-8s, 12-28s, 32-48s, and 52-58s), the disappearance of a ring and PEA signal when the former approaches a cell pole (8, 28 and 48s), and the subsequent assembly of a new ring at/near midcell concomitant with the appearance of a new PEA signal in between the new ring and the pole previously devoid of signal (12, 32, and 52 s). Arrowheads (< or >) indicate both the position of the ring as well as the direction of its movement.
Shown is a cell of strain HL1/pDR174 [DminDE/Plac::bfp-minD, minE-gfp]. Cells were grown at 30°C in minimal medium supplemented with 37mM IPTG, and displayed a normal division pattern. For additional details, see [3].
References
- Raskin, D. M., and de Boer, P. A. J. (1999a). MinDE dependent pole-to-pole oscillation of division inhibitor MinC in Escherichiacoli., J Bacteriol 181, 6419-6424.
- Raskin, D. M., and de Boer, P. A. J. (1999b). Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichiacoli., Proc Natl Acad Sci USA 96, 4971-4976.
- Hale, C. A., Meinhardt, H., and de Boer, P. A. J. (2001). Dynamic localization cycle of the cell division regulator MinE in E.coli., EMBO J. 20, 1563-1572.

