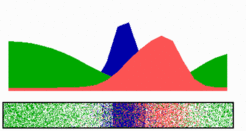
The MinCD pattern does not require prelocalized determinants and finds rapidly the new center after division
A membrane-associated patterning system (MinE, red) both depends on membrane-bound MinD (green), as well as causes its dissociation. In this way, a MinE maximum destabilizes itself, causing its shift towards a region of higher MinD concentration. On its way, the MinE wave removes MinD from the membrane. Shortly before the MinE wave reaches the pole, MinD and MinE concentrations collapse. Meanwhile, a new plateau of membrane-bound MinD is rising in the other part of the cell. A new high MinE concentration is triggered at its flank, causing this peak to disappear also, and so on. A system for septum formation is assumed to be also a pattern forming system (FtsZ, blue). An inhibition by MinD (green) is sufficient for a reliable localization of the FtsZ pattern in the center of the field. Note that in actual E.coli cells, the inhibition of FtsZ self-assembly is accomplished by the MinC protein which binds to, and co-oscillates with, MinD.

An important point in this model is that the center-finding mechanism does not require any prelocalizd components In the simulation above, the system is initiated by random fluctuations. After some irregular peaks flash up, the system enters its normal mode. Note that before normal oscillation is achieved, the FtsZ signal for initiation of division can emerge at a non-central position. It rapidly becomes positioned correctly, however, as soon as MinD oscillation starts (see also equation and parameters ).
The following simulation shows first the situation in a large cell. After division, the center is rapidly detected in the smaller cell: